$900K NSF grant helps WFU researchers look for the big picture in big data
Ever put an unripe avocado into a bag with a banana to make it guacamole-ready?
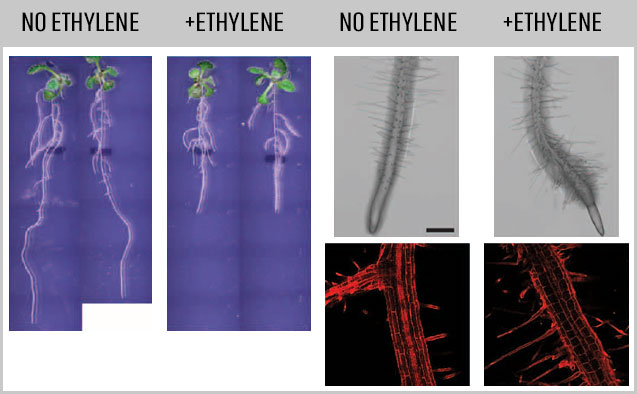
Ripening in the avocado is driven by the plant hormone ethylene, which is a gas that is released from fruit and other plant tissues. This important hormone controls many other aspects of plant development.
Now, thanks to a $900,000 award from the National Science Foundation, Wake Forest University researchers are examining how this hormone affects growth and development of the roots of Arabidopsis thaliana, which is a genetic model used to provide insight into other plants. Arabidopsis is used widely because it is short-lived, grows very quickly, is small in physical size, and has the best characterized genome of all plants.

This collaborative grant brings together researchers in biology, computer science and mathematics who began working together through Wake Forest’s Center for Molecular Signaling.
The Center brings together researchers from the life, physical, computational and mathematical sciences to identify how molecular signals facilitate communication between and within cells. An interdisciplinary team is needed for a systems biology approach, in which large data sets describing responses of all 22,000+ genes in the Arabidopsis genome change in response to this hormone. These large data sets, generated by the Wake Forest research team and other groups worldwide, are integrated to understand the global changes that remodel plant development.
“The funding will allow us to take these large data sets collected from prior experiments and look for patterns and relationships we could never see without the big data expertise of computer scientists and mathematicians,” said lead researcher Gloria Muday, professor of biology and director of the Center for Molecular Signaling. The computational and mathematical modeling approaches developed in this project will also be applicable in helping to identify patterns and networks in data sets from other organisms.
Traditionally, biologists who want to understand how processes are controlled at a molecular level look at changes in expression of one individual gene at a time.
“Our team will with work with groups of genes changing across a time course of ethylene treatment and apply new modeling approaches to unravel the complex signaling web. High levels of ethylene have a negative effect on root growth and development,” said Muday.
“Our work has important agricultural implications because root structure determines the amount of moisture and nutrients a plant takes from the soil. Ethylene has important roles in modulating plant growth under stressful environments.” Gloria Muday, professor of biology & dir. of the Center for Molecular Signaling
The research team includes biology professor Gloria Muday, computer science professors David John and William Turkett, math professor Jim Norris, and assistant research professor of biology Hanya Chrispeels. Collaborators from the University of Tennessee and Virginia Commonwealth University are also participants.
 The NSF grant also supports an ongoing outreach program called “Teaching with Tomatoes,” which uses plants with visibly striking mutations, including those with altered ethylene signaling, to teach genetics to high school students. Over the last four years, the program has taken more than 250 Wake Forest undergraduate biology students into secondary schools to lead case-study-based exercises for more than 1,000 students.
The NSF grant also supports an ongoing outreach program called “Teaching with Tomatoes,” which uses plants with visibly striking mutations, including those with altered ethylene signaling, to teach genetics to high school students. Over the last four years, the program has taken more than 250 Wake Forest undergraduate biology students into secondary schools to lead case-study-based exercises for more than 1,000 students.
Categories: Experiential Learning, Research & Discovery